DNA origami as a platform for colloidal self-assembly
Creating a generic patchy colloid has been a long sought-after Holy Grail in the field of colloidal assembly. We've achieved this goal using DNA origami to control the valency, dihedral angle and interaction strength at will. Starting with a 8064 circular ssDNA loop taken from bacteria called a scaffold, we use computer software to design about 200 shorter oligos, called staples which bind to and fold the scaffold into the desired shape using DNA hybridization. In this fashion, we program DNA molecules to self-assemble into colloids of 50 nm size with arbitrary shape to subnanometer accuracy, engineer the number and angle of the bonds to 2 degrees accuracy, and vary the inter-colloid interaction strength to a fraction of kT.
To make an icosahedral capsid, we pipetted 200 sequences of ssDNA into a test tube, heated it up to 65C and then while cooling down, the DNA strands self-assembled into structured colloids made
from 8064 basepairs of DNA. Upon further cooling, these colloids hierarchically self-assembled in the same tube into capsids containing over 160,000 basepairs of DNA, each positioned in
their designed location!
Our first platform for self-assembly is based on a triangular monomer building block. With this one basic design, countless variations are feasible to use to form a wide variety of self-assembled
structures. Significantly, the yield of the monomers and assembled structures is high, pointing to the utility of this approach for applications.
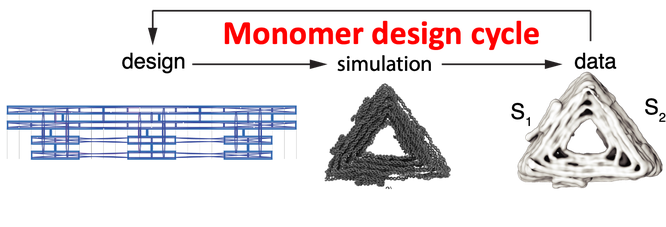
The success of applying DNA origami to colloidal self-assembly relies on a data-driven closed feedback loop between computer aided design, molecular dynamics simulation, theory and cryo-electronmicroscopy to guide the assembly of a colloidal monomer building block from several hundred strands of ssDNA.
Design principles for icosahedral capsids

Design principles for icosahedral shells. We built 8 colloidal patchy particles, shown in C, in which we controlled the valency, bond angle and interaction strength, which then self-assembled into 5 different capsids, shown in D. The octahedral (O), T=1 and T=3 capsids required one species of triangle monomer for assembly. T=4 required 2 distinct triangular monomers and T=9, the largest structure we built, required 3 distinct triangular monomers. A, Icosahedral shell encapsulating a virus capsid, an application of this technology. B, Triangular net representation of icosahedral shells forms the basis of the Caspar-Klug theory of virus structure. Each colored triangle represents one of the 20 faces forming an icosahedron. (h,k) indicates the location of the pentamers within a shell. C, Each cylinder represents dsDNA. Cylindrical models of DNA origami monomers in the shape of triangles that assemble into the shells shown in D. Each triangle is formed from a 8064 circular ssDNA loop (scaffold). About 200 shorter oligos (staples) were designed to fold the scaffold into the desired shape using DNA hybridization. The edges of the triangles are bevelled and modified with shape-complementary protrusions (light) and recesses (dark). The arrows indicate shape-complementary combinations. Triangles are bound together through blunt end interactions, not DNA hybridization. Subscripts (such as ‘pent’) on T refer to triangle names; S1 to S3 number the sides of each triangle. D, The octahedral and icosahedral shells formed by the triangles shown in C. For each shell design, one of the polyhedral faces has been displaced (see B) to help show the polyhedral symmetry. O, octahedral; T, T number; α, the bevel angle of the sides of the triangles; #, the number of DNA origami triangles building the shell.

(L to R) Mike Hagan, Don Caspar and Seth Fraden at the Rosenstiel awards in 2019. Don is thrilled to see his principle of quasi-equivalence brought to life with DNA origami triangles (pictured on the tablet).
T=1 Monomer
Cryo-EM reconstruction of T = 1 monomer triangle. An animation showing the docking of two monomers. Twenty monomers assemble into a closed icosahedron.
T=1 Monomer Slice
Cryo-EM reconstruction of T=1 monomer. The data set is three dimensional so it is possible to reveal the interior of the triangle. Notice the cross-overs as the sections proceed along the DNA double helix.
T=1 Capsid
Cryo-EM structure of assembled T = 1 icosahedron capsid. The 3D nature of cryo allows the electron density to be visualized in the interior of the capsid. Scale bar is 20 nm.
T=3 capsid design

Cryo-EM of T=3 monomer
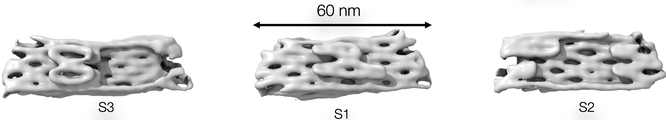
T=3 Monomer
Cryo-EM reconstruction of the T=3 monomer, which is an isosceles triangle with S1 = S2 = 52nm and S3 = 59nm. S3 binds to S3 to form the pseudo 6-fold vertex and S1 binds to S2 to form the 5-fold vertex. It was designed to have equal dihedral angles of 11.6 degrees and the measured angle is 13.7 degrees.
T=3 Capsid
Cryo-EM reconstruction of the T = 3 assembled capsid comprises 60 identical triangles. All Caspar-Klug capsids have 20T triangles, 12 5-fold vertices and 10(T -1) 6-fold vertices. The outer diameter is 200nm and the inner diameter is 160nm.
Encapsulation

The interior face of the origami monomer (triangle) is functionalized with 9 ssDNA "handles" that are complementary to ssDNA handles on the cargo. In this case the cargo is the 8064 circular ssDNA that serves as the scaffold of the monomer. So in some sense, we are encapsulating the origami "genome" inside the self-assembled origami capsid, recapitulating the action of an actual virus.
A TEM negative stain tomo-gram of a completed T=1 capsid encapsulating a bare circular 8064 base ssDNA. The inner diameter is 83 nm and the outer diameter is 114 nm.

A TEM negative stain tomo-gram of a completed T=1 capsid encapsulating a 30 nm gold sphere.

A TEM negative stain tomo-gram of a completed T=1 capsid encapsulating a circular 8064 base ssDNA that is labeled with 15 nm diameter gold spheres.

Anti-viral strategy

Our colleagues in the Dietz lab at TUM devised a strategy to prevent viruses from entering cells thereby thwarting infection. They modified the capsid design to make open structures and added antibodies to the bottom of the capsids. The concept is to trap the virus deep inside the cup-shaped origami frame so that the virus cannot touch the host's cell wall and thereby the virus is unable to trigger ruptureless transport across the cell membrane.
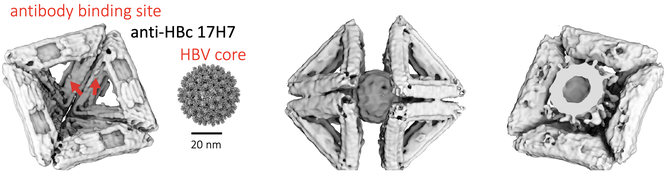

A half of an octahedron is functionalized with antibodies to Human Hepatitis B virus core. Two octahedral halves surround the virus, as anticipated by George Lucas in 1977.

Modified T=1 icosahedra are used to capture HBV shown in cryo and negative stain TEM.
Cylinders
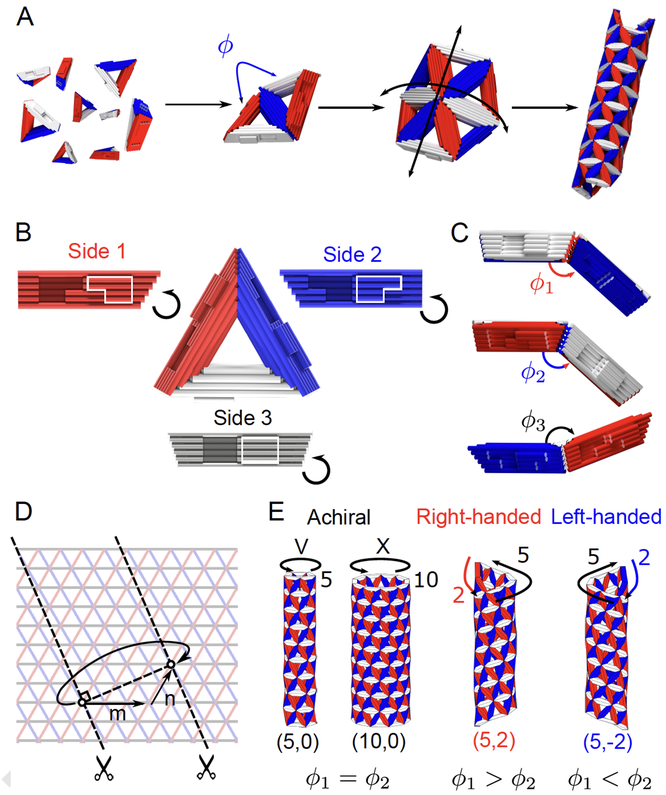
Cylinders differ from capsids in that the Gaussian curvature is zero for cylinders and positive for capsids. To build cylinders from triangles, two of the three dihedral angles are positive and one negative. The dihedral angles set the diameter and chirality of the cylinder. The binding rules for triangles that will assemble into cylinders are S1-S1, S2-S2 and S3-S3.
Cryo-EM reconstruction of a triangular monomer designed to assemble into a cylinder. The binding rules are S1-S1, S2-S2 and S3-S3. The electron density morphs onto the structure as calculated by OxDNA, a molecular dynamics package designed to simulate DNA origami.

TEM of assembled cylinder.
Data driven design cycles

The success of this application of DNA origami to colloidal self-assembly relies on a two nested data-driven closed feedback loops between computer aided design, molecular dynamics simulation, theory and cryo-electronmicroscopy. One is the design cycle for the self-assembly of the monomer building block from 200 oligos of ssDNA. The other design cycle is for the self-assembly of the colloidal building block into composite structures, e.g. capsids and tubules.
Support
This research was supported by the NSF Brandeis Bioinspired Soft Materials MRSEC - 2011846.
Publications
-
Programmable icosahedral shell system for virus trapping
Sigl, C., Willner, E. M., Engelen, W., Kretzmann, J. A., Sachenbacher, K., Liedl, A., Kolbe, F., Wilsch, F., Aghvami, S. A., Protzer, U., Hagan, M. F., Fraden, S. and Dietz, H.
Nature Materials, (2021) DOI: 10.1038/s41563-021-01020-4
Supplement. -
Geometrically programmed
self-limited assembly of tubules using DNA origami colloids
Daichi Hayakawa, Thomas E. Videbæk, Douglas M. Hall, Huang Fang, Christian Sigl, Elija Feigl, Hendrik Dietz, Seth Fraden, Michael F. Hagan, Gregory M. Grason, W. Benjamin Rogers
PNAS Vol. 119, e2207902119 (2022). DOI: 10.1073/pnas.2207902119.